LangChain’s Father or mother Doc Retriever — Revisited | by Omri Eliyahu Levy
TL;DR — We obtain the identical performance as LangChains’ Father or mother Doc Retriever (link) by using metadata queries. You may discover the code here.
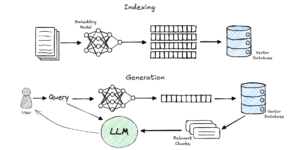
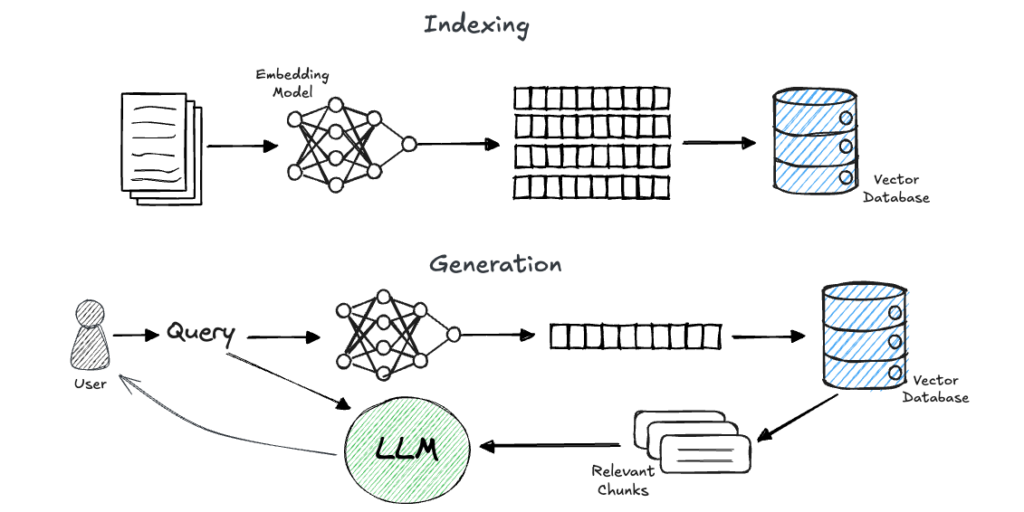
Retrieval-augmented technology (RAG) is at the moment one of many hottest matters on this planet of LLM and AI purposes.
In brief, RAG is a method for grounding a generative fashions’ response on chosen information sources. It includes two phases: retrieval and technology.
- Within the retrieval section, given a consumer’s question, we retrieve items of related info from a predefined information supply.
- Then, we insert the retrieved info into the immediate that’s despatched to an LLM, which (ideally) generates a solution to the consumer’s query primarily based on the supplied context.
A generally used method to realize environment friendly and correct retrieval is thru the utilization of embeddings. On this method, we preprocess customers’ knowledge (let’s assume plain textual content for simplicity) by splitting the paperwork into chunks (resembling pages, paragraphs, or sentences). We then use an embedding mannequin to create a significant, numerical illustration of those chunks, and retailer them in a vector database. Now, when a question is available in, we embed it as nicely and carry out a similarity search utilizing the vector database to retrieve the related info
If you’re fully new to this idea, I’d suggest deeplearning.ai nice course, LangChain: Chat with Your Data.
“Father or mother Doc Retrieval” or “Sentence Window Retrieval” as referred by others, is a standard method to reinforce the efficiency of retrieval strategies in RAG by offering the LLM with a broader context to think about.
In essence, we divide the unique paperwork into comparatively small chunks, embed each, and retailer them in a vector database. Utilizing such small chunks (a sentence or a few sentences) helps the embedding fashions to raised replicate their that means [1].
Then, at retrieval time, we don’t return probably the most comparable chunk as discovered by the vector database solely, but additionally its surrounding context (chunks) within the authentic doc. That approach, the LLM may have a broader context, which, in lots of instances, helps generate higher solutions.
LangChain helps this idea by way of Father or mother Doc Retriever [2]. The Father or mother Doc Retriever permits you to: (1) retrieve the complete doc a particular chunk originated from, or (2) pre-define a bigger “mum or dad” chunk, for every smaller chunk related to that mum or dad.
Let’s discover the instance from LangChains’ docs:
# This textual content splitter is used to create the mum or dad paperwork
parent_splitter = RecursiveCharacterTextSplitter(chunk_size=2000)
# This textual content splitter is used to create the kid paperwork
# It ought to create paperwork smaller than the mum or dad
child_splitter = RecursiveCharacterTextSplitter(chunk_size=400)
# The vectorstore to make use of to index the kid chunks
vectorstore = Chroma(
collection_name="split_parents", embedding_function=OpenAIEmbeddings()
)
# The storage layer for the mum or dad paperwork
retailer = InMemoryStore()
retriever = ParentDocumentRetriever(
vectorstore=vectorstore,
docstore=retailer,
child_splitter=child_splitter,
parent_splitter=parent_splitter,
)
retrieved_docs = retriever.invoke("justice breyer")
For my part, there are two disadvantages of the LangChains’ method:
- The necessity to handle exterior storage to learn from this convenient method, both in reminiscence or one other persistent retailer. In fact, for actual use instances, the InMemoryStore used within the varied examples won’t suffice.
- The “mum or dad” retrieval isn’t dynamic, that means we can not change the dimensions of the encompassing window on the fly.
Certainly, a couple of questions have been raised concerning this challenge [3].
Right here I’ll additionally point out that Llama-index has its personal SentenceWindowNodeParser [4], which typically has the identical disadvantages.
In what follows, I’ll current one other method to realize this convenient characteristic that addresses the 2 disadvantages talked about above. On this method, we’ll be solely utilizing the vector retailer that’s already in use.
Different Implementation
To be exact, we’ll be utilizing a vector retailer that helps the choice to carry out metadata queries solely, with none similarity search concerned. Right here, I’ll current an implementation for ChromaDB and Milvus. This idea might be simply tailored to any vector database with such capabilities. I’ll consult with Pinecone for instance ultimately of this tutorial.
The overall idea
The idea is simple:
- Development: Alongside every chunk, save in its metadata the document_id it was generated from and in addition the sequence_number of the chunk.
- Retrieval: After performing the same old similarity search (assuming for simplicity solely the highest 1 end result), we get hold of the document_id and the sequence_number of the chunk from the metadata of the retrieved chunk. Retrieve all chunks with surrounding sequence numbers which have the identical document_id.
For instance, assuming you’ve listed a doc named instance.pdf in 80 chunks. Then, for some question, you discover that the closest vector is the one with the next metadata:
{document_id: "instance.pdf", sequence_number: 20}
You may simply get all vectors from the identical doc with sequence numbers from 15 to 25.
Let’s see the code.
Right here, I’m utilizing:
chromadb==0.4.24
langchain==0.2.8
pymilvus==2.4.4
langchain-community==0.2.7
langchain-milvus==0.1.2
The one attention-grabbing factor to note beneath is the metadata related to every chunk, which is able to permit us to carry out the search.
from langchain_community.document_loaders import PyPDFLoader
from langchain_core.paperwork import Doc
from langchain_text_splitters import RecursiveCharacterTextSplitterdocument_id = "instance.pdf"
def preprocess_file(file_path: str) -> listing[Document]:
"""Load pdf file, chunk and construct applicable metadata"""
loader = PyPDFLoader(file_path=file_path)
pdf_docs = loader.load()
text_splitter = RecursiveCharacterTextSplitter(
chunk_size=1000,
chunk_overlap=0,
)
docs = text_splitter.split_documents(paperwork=pdf_docs)
chunks_metadata = [
{"document_id": file_path, "sequence_number": i} for i, _ in enumerate(docs)
]
for chunk, metadata in zip(docs, chunks_metadata):
chunk.metadata = metadata
return docs
Now, lets implement the precise retrieval in Milvus and Chroma. Be aware that I’ll use the LangChains’ objects and never the native shoppers. I do that as a result of I assume builders would possibly wish to preserve LangChains’ helpful abstraction. Alternatively, it is going to require us to carry out some minor hacks to bypass these abstractions in a database-specific approach, so you need to take that into consideration. Anyway, the idea stays the identical.
Once more, let’s assume for simplicity we would like solely probably the most comparable vector (“prime 1”). Subsequent, we’ll extract the related document_id and its sequence quantity. This can permit us to retrieve the encompassing window.
from langchain_community.vectorstores import Milvus, Chroma
from langchain_community.embeddings import DeterministicFakeEmbeddingembedding = DeterministicFakeEmbedding(measurement=384) # Only for the demo :)
def parent_document_retrieval(
question: str, consumer: Milvus | Chroma, window_size: int = 4
):
top_1 = consumer.similarity_search(question=question, okay=1)[0]
doc_id = top_1.metadata["document_id"]
seq_num = top_1.metadata["sequence_number"]
ids_window = [seq_num + i for i in range(-window_size, window_size, 1)]
# ...
Now, for the window/mum or dad retrieval, we’ll dig beneath the Langchain abstraction, in a database-specific approach.
For Milvus:
if isinstance(consumer, Milvus):
expr = f"document_id LIKE '{doc_id}' && sequence_number in {ids_window}"
res = consumer.col.question(
expr=expr, output_fields=["sequence_number", "text"], restrict=len(ids_window)
) # That is Milvus particular
docs_to_return = [
Document(
page_content=d["text"],
metadata={
"sequence_number": d["sequence_number"],
"document_id": doc_id,
},
)
for d in res
]
# ...
For Chroma:
elif isinstance(consumer, Chroma):
expr = {
"$and": [
{"document_id": {"$eq": doc_id}},
{"sequence_number": {"$gte": ids_window[0]}},
{"sequence_number": {"$lte": ids_window[-1]}},
]
}
res = consumer.get(the place=expr) # That is Chroma particular
texts, metadatas = res["documents"], res["metadatas"]
docs_to_return = [
Document(
page_content=t,
metadata={
"sequence_number": m["sequence_number"],
"document_id": doc_id,
},
)
for t, m in zip(texts, metadatas)
]
and don’t overlook to type it by the sequence quantity:
docs_to_return.type(key=lambda x: x.metadata["sequence_number"])
return docs_to_return
In your comfort, you may discover the complete code here.
Pinecone (and others)
So far as I do know, there’s no native solution to carry out such a metadata question in Pinecone, however you may natively fetch vectors by their ID (https://docs.pinecone.io/guides/data/fetch-data).
Therefore, we are able to do the next: every chunk will get a novel ID, which is actually a concatenation of the document_id and the sequence quantity. Then, given a vector retrieved within the similarity search, you may dynamically create an inventory of the IDs of the encompassing chunks and obtain the identical end result.
It’s value mentioning that vector databases weren’t designed to carry out “common” database operations and normally not optimized for that, and every database will carry out in a different way. Milvus, for instance, will assist constructing indices over scalar fields (“metadata”) which may optimize these sorts of queries.
Additionally, notice that it requires further question to the vector database. First we retrieved probably the most comparable vector, after which we carried out further question to get the encompassing chunks within the authentic doc.
And naturally, as seen from the code examples above, the implementation is vector database-specific and isn’t supported natively by the LangChains’ abstraction.
On this weblog we launched an implementation to realize sentence-window retrieval, which is a helpful retrieval approach utilized in many RAG purposes. On this implementation we’ve used solely the vector database which is already in use anyway, and in addition assist the choice to switch dynamically the the dimensions of the encompassing window retrieved.
[1] ARAGOG: Superior RAG Output Grading, https://arxiv.org/pdf/2404.01037, part 4.2.2
[2] https://python.langchain.com/v0.1/docs/modules/data_connection/retrievers/parent_document_retriever/
[3] Some associated points:
– https://github.com/langchain-ai/langchain/issues/14267
– https://github.com/langchain-ai/langchain/issues/20315
– https://stackoverflow.com/questions/77385587/persist-parentdocumentretriever-of-langchain
[4] https://docs.llamaindex.ai/en/stable/api_reference/node_parsers/sentence_window/