Construct a medical imaging AI inference pipeline with MONAI Deploy on AWS
This put up is cowritten with Ming (Melvin) Qin, David Bericat and Brad Genereaux from NVIDIA.
Medical imaging AI researchers and builders want a scalable, enterprise framework to construct, deploy, and combine their AI purposes. AWS and NVIDIA have come collectively to make this imaginative and prescient a actuality. AWS, NVIDIA, and other partners construct purposes and options to make healthcare extra accessible, inexpensive, and environment friendly by accelerating cloud connectivity of enterprise imaging. MONAI Deploy is without doubt one of the key modules inside MONAI (Medical Open Community for Synthetic Intelligence) developed by a consortium of educational and business leaders, together with NVIDIA. AWS HealthImaging (AHI) is a HIPAA-eligible, extremely scalable, performant, and cost-effective medical imagery retailer. We have now developed a MONAI Deploy connector to AHI to combine medical imaging AI purposes with subsecond picture retrieval latencies at scale powered by cloud-native APIs. The MONAI AI fashions and purposes may be hosted on Amazon SageMaker, which is a totally managed service to deploy machine studying (ML) fashions at scale. SageMaker takes care of organising and managing situations for inference and offers built-in metrics and logs for endpoints that you should use to observe and obtain alerts. It additionally affords a wide range of NVIDIA GPU instances for ML inference, in addition to a number of mannequin deployment choices with automated scaling, together with real-time inference, serverless inference, asynchronous inference, and batch transform.
On this put up, we show tips on how to deploy a MONAI Software Package deal (MAP) with the connector to AWS HealthImaging, utilizing a SageMaker multi-model endpoint for real-time inference and asynchronous inference. These two choices cowl a majority of near-real-time medical imaging inference pipeline use instances.
Answer overview
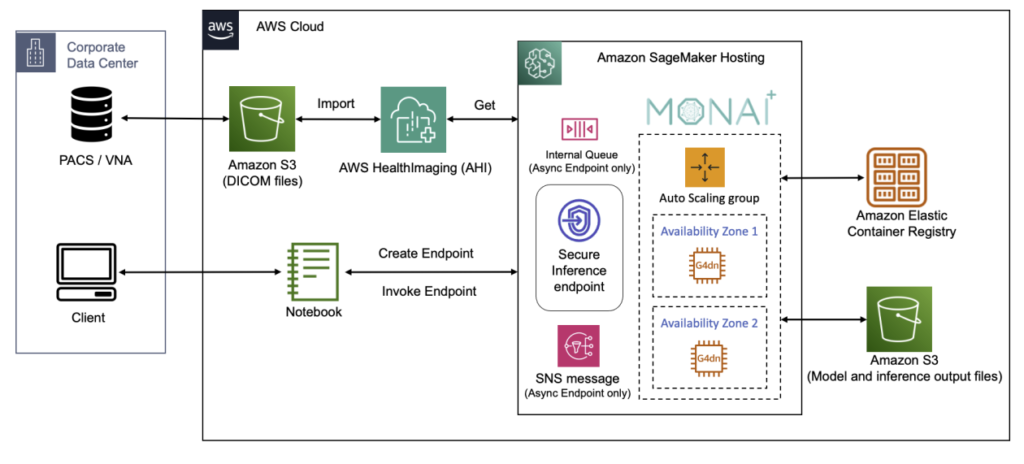
The next diagram illustrates the answer structure.
Stipulations
Full the next prerequisite steps:
- Use an AWS account with one of many following Areas, the place AWS HealthImaging is offered: North Virginia (
us-east-1), Oregon (us-west-2), Eire (eu-west-1), and Sydney (ap-southeast-2). - Create an Amazon SageMaker Studio domain and user profile with AWS Identity and Access Management (IAM) permission to entry AWS HealthImaging.
- Enable the JupyterLab v3 extension and set up Imjoy-jupyter-extension if you wish to visualize medical photos on SageMaker pocket book interactively utilizing itkwidgets.
MAP connector to AWS HealthImaging
AWS HealthImaging imports DICOM P10 recordsdata and converts them into ImageSets, that are a optimized illustration of a DICOM collection. AHI offers API entry to ImageSet metadata and ImageFrames. Metadata incorporates all DICOM attributes in a JSON doc. ImageFrames are returned encoded within the High-Throughput JPEG2000 (HTJ2K) lossless format, which may be decoded extraordinarily quick. ImageSets may be retrieved through the use of the AWS Command Line Interface (AWS CLI) or the AWS SDKs.
MONAI is a medical imaging AI framework that takes analysis breakthroughs and AI purposes into scientific affect. MONAI Deploy is the processing pipeline that permits the end-to-end workflow, together with packaging, testing, deploying, and operating medical imaging AI purposes in scientific manufacturing. It includes the MONAI Deploy App SDK, MONAI Deploy Express, Workflow Manager, and Informatics Gateway. The MONAI Deploy App SDK offers ready-to-use algorithms and a framework to speed up constructing medical imaging AI purposes, in addition to utility instruments to bundle the applying right into a MAP container. The built-in standards-based functionalities within the app SDK permit the MAP to easily combine into well being IT networks, which requires the usage of requirements reminiscent of DICOM, HL7, and FHIR, and throughout information heart and cloud environments. MAPs can use each predefined and customised operators for DICOM picture loading, collection choice, mannequin inference, and postprocessing
We have now developed a Python module utilizing the AWS HealthImaging Python SDK Boto3. You’ll be able to pip set up it and use the helper operate to retrieve DICOM Service-Object Pair (SOP) situations as follows:
!pip set up -q AHItoDICOMInterface
from AHItoDICOMInterface.AHItoDICOM import AHItoDICOM
helper = AHItoDICOM()
situations = helper.DICOMizeImageSet(datastore_id=datastoreId , image_set_id=subsequent(iter(imageSetIds)))The output SOP situations may be visualized utilizing the interactive 3D medical picture viewer itkwidgets within the following notebook. The AHItoDICOM class takes benefit of a number of processes to retrieve pixel frames from AWS HealthImaging in parallel, and decode the HTJ2K binary blobs utilizing the Python OpenJPEG library. The ImageSetIds come from the output recordsdata of a given AWS HealthImaging import job. Given the DatastoreId and import JobId, you’ll be able to retrieve the ImageSetId, which is equal to the DICOM collection occasion UID, as follows:
imageSetIds = {}
strive:
response = s3.head_object(Bucket=OutputBucketName, Key=f"output/{res_createstore['datastoreId']}-DicomImport-{res_startimportjob['jobId']}/job-output-manifest.json")
if response['ResponseMetadata']['HTTPStatusCode'] == 200:
information = s3.get_object(Bucket=OutputBucketName, Key=f"output/{res_createstore['datastoreId']}-DicomImport-{res_startimportjob['jobId']}/SUCCESS/success.ndjson")
contents = information['Body'].learn().decode("utf-8")
for l in contents.splitlines():
isid = json.hundreds(l)['importResponse']['imageSetId']
if isid in imageSetIds:
imageSetIds[isid]+=1
else:
imageSetIds[isid]=1
besides ClientError:
moveWith ImageSetId, you’ll be able to retrieve the DICOM header metadata and picture pixels individually utilizing native AWS HealthImaging API capabilities. The DICOM exporter aggregates the DICOM headers and picture pixels into the Pydicom dataset, which may be processed by the MAP DICOM data loader operator. Utilizing the DICOMizeImageSet()operate, we now have created a connector to load picture information from AWS HealthImaging, primarily based on the MAP DICOM data loader operator:
class AHIDataLoaderOperator(Operator):
def __init__(self, ahi_client, must_load: bool = True, *args, **kwargs):
self.ahi_client = ahi_client
…
def _load_data(self, input_obj: string):
study_dict = {}
series_dict = {}
sop_instances = self.ahi_client.DICOMizeImageSet(input_obj['datastoreId'], input_obj['imageSetId'])Within the previous code, ahi_client is an occasion of the AHItoDICOM DICOM exporter class, with information retrieval capabilities illustrated. We have now included this new information loader operator right into a 3D spleen segmentation AI application created by the MONAI Deploy App SDK. You’ll be able to first discover tips on how to create and run this software on a local notebook instance, after which deploy this MAP software into SageMaker managed inference endpoints.
SageMaker asynchronous inference
A SageMaker asynchronous inference endpoint is used for requests with massive payload sizes (as much as 1 GB), lengthy processing occasions (as much as quarter-hour), and near-real-time latency necessities. When there are not any requests to course of, this deployment choice can downscale the occasion depend to zero for value financial savings, which is good for medical imaging ML inference workloads. Comply with the steps within the sample notebook to create and invoke the SageMaker asynchronous inference endpoint. To create an asynchronous inference endpoint, you will have to create a SageMaker mannequin and endpoint configuration first. To create a SageMaker mannequin, you will have to load a mannequin.tar.gz bundle with a defined directory structure right into a Docker container. The mannequin.tar.gz bundle features a pre-trained spleen segmentation mannequin.ts file and a personalized inference.py file. We have now used a prebuilt container with Python 3.8 and PyTorch 1.12.1 framework variations to load the mannequin and run predictions.
Within the personalized inference.py file, we instantiate an AHItoDICOM helper class from AHItoDICOMInterface and use it to create a MAP occasion within the model_fn() operate, and we run the MAP software on each inference request within the predict_fn() operate:
from app import AISpleenSegApp
from AHItoDICOMInterface.AHItoDICOM import AHItoDICOM
helper = AHItoDICOM()
def model_fn(model_dir, context):
…
monai_app_instance = AISpleenSegApp(helper, do_run=False,path="/residence/model-server")
def predict_fn(input_data, mannequin):
with open('/residence/model-server/inputImageSets.json', 'w') as f:
f.write(json.dumps(input_data))
output_folder = "/residence/model-server/output"
if not os.path.exists(output_folder):
os.makedirs(output_folder)
mannequin.run(enter="/residence/model-server/inputImageSets.json", output=output_folder, workdir="/residence/model-server", mannequin="/decide/ml/mannequin/mannequin.ts")To invoke the asynchronous endpoint, you will have to add the request enter payload to Amazon Simple Storage Service (Amazon S3), which is a JSON file specifying the AWS HealthImaging datastore ID and ImageSet ID to run inference on:
sess = sagemaker.Session()
InputLocation = sess.upload_data('inputImageSets.json', bucket=sess.default_bucket(), key_prefix=prefix, extra_args={"ContentType": "software/json"})
response = runtime_sm_client.invoke_endpoint_async(EndpointName=endpoint_name, InputLocation=InputLocation, ContentType="software/json", Settle for="software/json")
output_location = response["OutputLocation"]The output may be present in Amazon S3 as properly.
SageMaker multi-model real-time inference
SageMaker real-time inference endpoints meet interactive, low-latency necessities. This selection can host a number of fashions in a single container behind one endpoint, which is a scalable and cost-effective resolution to deploying a number of ML fashions. A SageMaker multi-model endpoint uses NVIDIA Triton Inference Server with GPU to run a number of deep studying mannequin inferences.
On this part, we stroll by means of tips on how to create and invoke a multi-model endpoint adapting your own inference container within the following sample notebook. Completely different fashions may be served in a shared container on the identical fleet of assets. Multi-model endpoints cut back deployment overhead and scale mannequin inferences primarily based on the visitors patterns to the endpoint. We used AWS developer tools together with Amazon CodeCommit, Amazon CodeBuild, and Amazon CodePipeline to construct the customized container for SageMaker mannequin inference. We ready a model_handler.py to deliver your individual container as a substitute of the inference.py file within the earlier instance, and carried out the initialize(), preprocess(), and inference() capabilities:
from app import AISpleenSegApp
from AHItoDICOMInterface.AHItoDICOM import AHItoDICOM
class ModelHandler(object):
def __init__(self):
self.initialized = False
self.shapes = None
def initialize(self, context):
self.initialized = True
properties = context.system_properties
model_dir = properties.get("model_dir")
gpu_id = properties.get("gpu_id")
helper = AHItoDICOM()
self.monai_app_instance = AISpleenSegApp(helper, do_run=False, path="/residence/model-server/")
def preprocess(self, request):
inputStr = request[0].get("physique").decode('UTF8')
datastoreId = json.hundreds(inputStr)['inputs'][0]['datastoreId']
imageSetId = json.hundreds(inputStr)['inputs'][0]['imageSetId']
with open('/tmp/inputImageSets.json', 'w') as f:
f.write(json.dumps({"datastoreId": datastoreId, "imageSetId": imageSetId}))
return '/tmp/inputImageSets.json'
def inference(self, model_input):
self.monai_app_instance.run(enter=model_input, output="/residence/model-server/output/", workdir="/residence/model-server/", mannequin=os.environ["model_dir"]+"/mannequin.ts")After the container is constructed and pushed to Amazon Elastic Container Registry (Amazon ECR), you’ll be able to create SageMaker mannequin with it, plus completely different mannequin packages (tar.gz recordsdata) in a given Amazon S3 path:
model_name = "DEMO-MONAIDeployModel" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
model_url = "s3://{}/{}/".format(bucket, prefix)
container = "{}.dkr.ecr.{}.amazonaws.com/{}:dev".format( account_id, area, prefix )
container = {"Picture": container, "ModelDataUrl": model_url, "Mode": "MultiModel"}
create_model_response = sm_client.create_model(ModelName=model_name, ExecutionRoleArn=position, PrimaryContainer=container)It’s noteworthy that the model_url right here solely specifies the trail to a folder of tar.gz recordsdata, and also you specify which mannequin bundle to make use of for inference once you invoke the endpoint, as proven within the following code:
Payload = {"inputs": [ {"datastoreId": datastoreId, "imageSetId": next(iter(imageSetIds))} ]}
response = runtime_sm_client.invoke_endpoint(EndpointName=endpoint_name, ContentType="software/json", Settle for="software/json", TargetModel="mannequin.tar.gz", Physique=json.dumps(Payload))We will add extra fashions to the prevailing multi-model inference endpoint with out having to replace the endpoint or create a brand new one.
Clear up
Don’t overlook to finish the Delete the internet hosting assets step within the lab-3 and lab-4 notebooks to delete the SageMaker inference endpoints. You must flip down the SageMaker pocket book occasion to avoid wasting prices as properly. Lastly, you’ll be able to both name the AWS HealthImaging API operate or use the AWS HealthImaging console to delete the picture units and information retailer created earlier:
for s in imageSetIds.keys():
medicalimaging.deleteImageSet(datastoreId, s)
medicalimaging.deleteDatastore(datastoreId)Conclusion
On this put up, we confirmed you tips on how to create a MAP connector to AWS HealthImaging, which is reusable in purposes constructed with the MONAI Deploy App SDK, to combine with and speed up picture information retrieval from a cloud-native DICOM retailer to medical imaging AI workloads. The MONAI Deploy SDK can be utilized to assist hospital operations. We additionally demonstrated two internet hosting choices to deploy MAP AI purposes on SageMaker at scale.
Undergo the instance notebooks within the GitHub repository to be taught extra about tips on how to deploy MONAI purposes on SageMaker with medical photos saved in AWS HealthImaging. To know what AWS can do for you, contact an AWS representative.
For added assets, confer with the next:
In regards to the Authors
 Ming (Melvin) Qin is an impartial contributor on the Healthcare workforce at NVIDIA, centered on creating an AI inference software framework and platform to deliver AI to medical imaging workflows. Earlier than becoming a member of NVIDIA in 2018 as a founding member of Clara, Ming spent 15 years creating Radiology PACS and Workflow SaaS as lead engineer/architect at Stentor Inc., later acquired by Philips Healthcare to kind its Enterprise Imaging.
Ming (Melvin) Qin is an impartial contributor on the Healthcare workforce at NVIDIA, centered on creating an AI inference software framework and platform to deliver AI to medical imaging workflows. Earlier than becoming a member of NVIDIA in 2018 as a founding member of Clara, Ming spent 15 years creating Radiology PACS and Workflow SaaS as lead engineer/architect at Stentor Inc., later acquired by Philips Healthcare to kind its Enterprise Imaging.
 David Bericat is a product supervisor for Healthcare at NVIDIA, the place he leads the Undertaking MONAI Deploy working group to deliver AI from analysis to scientific deployments. His ardour is to speed up well being innovation globally translating it to true scientific affect. Beforehand, David labored at Crimson Hat, implementing open supply ideas on the intersection of AI, cloud, edge computing, and IoT. His proudest moments embrace mountaineering to the Everest base camp and enjoying soccer for over 20 years.
David Bericat is a product supervisor for Healthcare at NVIDIA, the place he leads the Undertaking MONAI Deploy working group to deliver AI from analysis to scientific deployments. His ardour is to speed up well being innovation globally translating it to true scientific affect. Beforehand, David labored at Crimson Hat, implementing open supply ideas on the intersection of AI, cloud, edge computing, and IoT. His proudest moments embrace mountaineering to the Everest base camp and enjoying soccer for over 20 years.
 Brad Genereaux is International Lead, Healthcare Alliances at NVIDIA, the place he’s answerable for developer relations with a spotlight in medical imaging to speed up synthetic intelligence and deep studying, visualization, virtualization, and analytics options. Brad evangelizes the ever present adoption and integration of seamless healthcare and medical imaging workflows into on a regular basis scientific observe, with greater than 20 years of expertise in healthcare IT.
Brad Genereaux is International Lead, Healthcare Alliances at NVIDIA, the place he’s answerable for developer relations with a spotlight in medical imaging to speed up synthetic intelligence and deep studying, visualization, virtualization, and analytics options. Brad evangelizes the ever present adoption and integration of seamless healthcare and medical imaging workflows into on a regular basis scientific observe, with greater than 20 years of expertise in healthcare IT.
 Gang Fu is a Healthcare Options Architect at AWS. He holds a PhD in Pharmaceutical Science from the College of Mississippi and has over 10 years of know-how and biomedical analysis expertise. He’s keen about know-how and the affect it might make on healthcare.
Gang Fu is a Healthcare Options Architect at AWS. He holds a PhD in Pharmaceutical Science from the College of Mississippi and has over 10 years of know-how and biomedical analysis expertise. He’s keen about know-how and the affect it might make on healthcare.
 JP Leger is a Senior Options Architect supporting tutorial medical facilities and medical imaging workflows at AWS. He has over 20 years of experience in software program engineering, healthcare IT, and medical imaging, with in depth expertise architecting techniques for efficiency, scalability, and safety in distributed deployments of enormous information volumes on premises, within the cloud, and hybrid with analytics and AI.
JP Leger is a Senior Options Architect supporting tutorial medical facilities and medical imaging workflows at AWS. He has over 20 years of experience in software program engineering, healthcare IT, and medical imaging, with in depth expertise architecting techniques for efficiency, scalability, and safety in distributed deployments of enormous information volumes on premises, within the cloud, and hybrid with analytics and AI.
 Chris Hafey is a Principal Options Architect at Amazon Internet Companies. He has over 25 years’ expertise within the medical imaging business and makes a speciality of constructing scalable high-performance techniques. He’s the creator of the favored CornerstoneJS open supply undertaking, which powers the favored OHIF open supply zero footprint viewer. He contributed to the DICOMweb specification and continues to work in direction of bettering its efficiency for web-based viewing.
Chris Hafey is a Principal Options Architect at Amazon Internet Companies. He has over 25 years’ expertise within the medical imaging business and makes a speciality of constructing scalable high-performance techniques. He’s the creator of the favored CornerstoneJS open supply undertaking, which powers the favored OHIF open supply zero footprint viewer. He contributed to the DICOMweb specification and continues to work in direction of bettering its efficiency for web-based viewing.