Effectively fine-tune the ESM-2 protein language mannequin with Amazon SageMaker
On this publish, we exhibit the way to effectively fine-tune a state-of-the-art protein language mannequin (pLM) to foretell protein subcellular localization utilizing Amazon SageMaker.
Proteins are the molecular machines of the physique, accountable for the whole lot from transferring your muscle groups to responding to infections. Regardless of this selection, all proteins are product of repeating chains of molecules referred to as amino acids. The human genome encodes 20 customary amino acids, every with a barely completely different chemical construction. These could be represented by letters of the alphabet, which then permits us to research and discover proteins as a textual content string. The large doable variety of protein sequences and constructions is what offers proteins their large number of makes use of.

Proteins additionally play a key function in drug improvement, as potential targets but in addition as therapeutics. As proven within the following desk, most of the top-selling medicine in 2022 had been both proteins (particularly antibodies) or different molecules like mRNA translated into proteins within the physique. Due to this, many life science researchers must reply questions on proteins sooner, cheaper, and extra precisely.
| Identify | Producer | 2022 International Gross sales ($ billions USD) | Indications |
| Comirnaty | Pfizer/BioNTech | $40.8 | COVID-19 |
| Spikevax | Moderna | $21.8 | COVID-19 |
| Humira | AbbVie | $21.6 | Arthritis, Crohn’s illness, and others |
| Keytruda | Merck | $21.0 | Varied cancers |
Knowledge supply: Urquhart, L. Top companies and drugs by sales in 2022. Nature Opinions Drug Discovery 22, 260–260 (2023).
As a result of we are able to characterize proteins as sequences of characters, we are able to analyze them utilizing methods initially developed for written language. This consists of giant language fashions (LLMs) pretrained on large datasets, which may then be tailored for particular duties, like textual content summarization or chatbots. Equally, pLMs are pre-trained on giant protein sequence databases utilizing unlabeled, self-supervised studying. We are able to adapt them to foretell issues just like the 3D construction of a protein or the way it might work together with different molecules. Researchers have even used pLMs to design novel proteins from scratch. These instruments don’t change human scientific experience, however they’ve the potential to hurry up pre-clinical improvement and trial design.
One problem with these fashions is their measurement. Each LLMs and pLMs have grown by orders of magnitude up to now few years, as illustrated within the following determine. Which means it could possibly take a very long time to coach them to enough accuracy. It additionally implies that you should use {hardware}, particularly GPUs, with giant quantities of reminiscence to retailer the mannequin parameters.

Lengthy coaching instances, plus giant situations, equals excessive value, which may put this work out of attain for a lot of researchers. For instance, in 2023, a research team described coaching a 100 billion-parameter pLM on 768 A100 GPUs for 164 days! Fortuitously, in lots of circumstances we are able to save time and assets by adapting an present pLM to our particular process. This method known as fine-tuning, and likewise permits us to borrow superior instruments from different kinds of language modeling.
Answer overview
The particular drawback we tackle on this publish is subcellular localization: Given a protein sequence, can we construct a mannequin that may predict if it lives on the surface (cell membrane) or within a cell? This is a vital piece of knowledge that may assist us perceive the perform and whether or not it will make a very good drug goal.
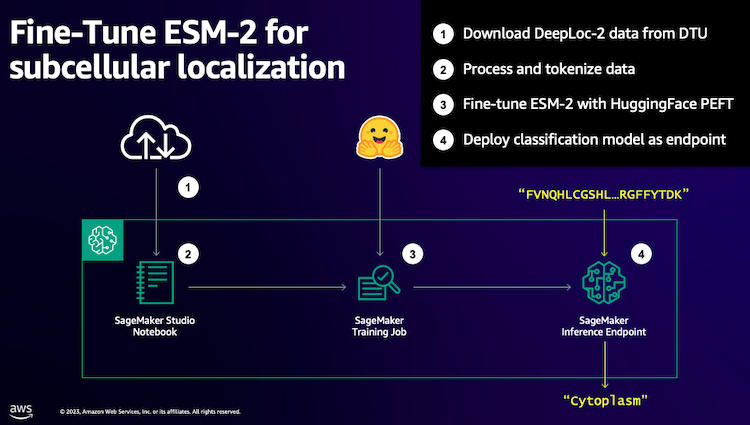
We begin by downloading a public dataset utilizing Amazon SageMaker Studio. Then we use SageMaker to fine-tune the ESM-2 protein language mannequin utilizing an environment friendly coaching methodology. Lastly, we deploy the mannequin as a real-time inference endpoint and use it to check some recognized proteins. The next diagram illustrates this workflow.

Within the following sections, we undergo the steps to arrange your coaching information, create a coaching script, and run a SageMaker coaching job. The entire code featured on this publish is accessible on GitHub.
Put together the coaching information
We use a part of the DeepLoc-2 dataset, which incorporates a number of thousand SwissProt proteins with experimentally decided areas. We filter for high-quality sequences between 100–512 amino acids:
df = pd.read_csv(
"https://companies.healthtech.dtu.dk/companies/DeepLoc-2.0/information/Swissprot_Train_Validation_dataset.csv"
).drop(["Unnamed: 0", "Partition"], axis=1)
df["Membrane"] = df["Membrane"].astype("int32")
# filter for sequences between 100 and 512 amino acides
df = df[df["Sequence"].apply(lambda x: len(x)).between(100, 512)]
# Take away pointless options
df = df[["Sequence", "Kingdom", "Membrane"]]
Subsequent, we tokenize the sequences and cut up them into coaching and analysis units:
dataset = Dataset.from_pandas(df).train_test_split(test_size=0.2, shuffle=True)
tokenizer = AutoTokenizer.from_pretrained("fb/esm2_t33_650M_UR50D")
def preprocess_data(examples, max_length=512):
textual content = examples["Sequence"]
encoding = tokenizer(textual content, truncation=True, max_length=max_length)
encoding["labels"] = examples["Membrane"]
return encoding
encoded_dataset = dataset.map(
preprocess_data,
batched=True,
num_proc=os.cpu_count(),
remove_columns=dataset["train"].column_names,
)
encoded_dataset.set_format("torch")
Lastly, we add the processed coaching and analysis information to Amazon Simple Storage Service (Amazon S3):
train_s3_uri = S3_PATH + "/information/prepare"
test_s3_uri = S3_PATH + "/information/take a look at"
encoded_dataset["train"].save_to_disk(train_s3_uri)
encoded_dataset["test"].save_to_disk(test_s3_uri)Create a coaching script
SageMaker script mode means that you can run your customized coaching code in optimized machine studying (ML) framework containers managed by AWS. For this instance, we adapt an existing script for text classification from Hugging Face. This permits us to strive a number of strategies for bettering the effectivity of our coaching job.
Methodology 1: Weighted coaching class
Like many organic datasets, the DeepLoc information is erratically distributed, that means there isn’t an equal variety of membrane and non-membrane proteins. We may resample our information and discard information from the bulk class. Nevertheless, this would scale back the entire coaching information and probably damage our accuracy. As a substitute, we calculate the category weights throughout the coaching job and use them to regulate the loss.
In our coaching script, we subclass the Coach class from transformers with a WeightedTrainer class that takes class weights into consideration when calculating cross-entropy loss. This helps stop bias in our mannequin:
class WeightedTrainer(Coach):
def __init__(self, class_weights, *args, **kwargs):
self.class_weights = class_weights
tremendous().__init__(*args, **kwargs)
def compute_loss(self, mannequin, inputs, return_outputs=False):
labels = inputs.pop("labels")
outputs = mannequin(**inputs)
logits = outputs.get("logits")
loss_fct = torch.nn.CrossEntropyLoss(
weight=torch.tensor(self.class_weights, system=mannequin.system)
)
loss = loss_fct(logits.view(-1, self.mannequin.config.num_labels), labels.view(-1))
return (loss, outputs) if return_outputs else lossMethodology 2: Gradient accumulation
Gradient accumulation is a coaching approach that enables fashions to simulate coaching on bigger batch sizes. Sometimes, the batch measurement (the variety of samples used to calculate the gradient in a single coaching step) is proscribed by the GPU reminiscence capability. With gradient accumulation, the mannequin calculates gradients on smaller batches first. Then, as a substitute of updating the mannequin weights immediately, the gradients get gathered over a number of small batches. When the gathered gradients equal the goal bigger batch measurement, the optimization step is carried out to replace the mannequin. This lets fashions prepare with successfully greater batches with out exceeding the GPU reminiscence restrict.
Nevertheless, additional computation is required for the smaller batch ahead and backward passes. Elevated batch sizes by way of gradient accumulation can decelerate coaching, particularly if too many accumulation steps are used. The intention is to maximise GPU utilization however keep away from extreme slowdowns from too many additional gradient computation steps.
Methodology 3: Gradient checkpointing
Gradient checkpointing is a way that reduces the reminiscence wanted throughout coaching whereas preserving the computational time affordable. Massive neural networks take up plenty of reminiscence as a result of they must retailer all of the intermediate values from the ahead cross with the intention to calculate the gradients throughout the backward cross. This will trigger reminiscence points. One resolution is to not retailer these intermediate values, however then they must be recalculated throughout the backward cross, which takes plenty of time.
Gradient checkpointing supplies a balanced strategy. It saves solely among the intermediate values, referred to as checkpoints, and recalculates the others as wanted. Due to this fact, it makes use of much less reminiscence than storing the whole lot, but in addition much less computation than recalculating the whole lot. By strategically deciding on which activations to checkpoint, gradient checkpointing allows giant neural networks to be educated with manageable reminiscence utilization and computation time. This necessary approach makes it possible to coach very giant fashions that will in any other case run into reminiscence limitations.
In our coaching script, we activate gradient activation and checkpointing by including the mandatory parameters to the TrainingArguments object:
from transformers import TrainingArguments
training_args = TrainingArguments(
gradient_accumulation_steps=4,
gradient_checkpointing=True
)Methodology 4: Low-Rank Adaptation of LLMs
Massive language fashions like ESM-2 can include billions of parameters which might be costly to coach and run. Researchers developed a coaching methodology referred to as Low-Rank Adaptation (LoRA) to make fine-tuning these large fashions extra environment friendly.
The important thing concept behind LoRA is that when fine-tuning a mannequin for a particular process, you don’t must replace all the unique parameters. As a substitute, LoRA provides new smaller matrices to the mannequin that remodel the inputs and outputs. Solely these smaller matrices are up to date throughout fine-tuning, which is way sooner and makes use of much less reminiscence. The unique mannequin parameters keep frozen.
After fine-tuning with LoRA, you may merge the small tailored matrices again into the unique mannequin. Or you may preserve them separate if you wish to rapidly fine-tune the mannequin for different duties with out forgetting earlier ones. General, LoRA permits LLMs to be effectively tailored to new duties at a fraction of the standard value.
In our coaching script, we configure LoRA utilizing the PEFT library from Hugging Face:
from peft import get_peft_model, LoraConfig, TaskType
import torch
from transformers import EsmForSequenceClassification
mannequin = EsmForSequenceClassification.from_pretrained(
“fb/esm2_t33_650M_UR50D”,
Torch_dtype=torch.bfloat16,
Num_labels=2,
)
peft_config = LoraConfig(
task_type=TaskType.SEQ_CLS,
inference_mode=False,
bias="none",
r=8,
lora_alpha=16,
lora_dropout=0.05,
target_modules=[
"query",
"key",
"value",
"EsmSelfOutput.dense",
"EsmIntermediate.dense",
"EsmOutput.dense",
"EsmContactPredictionHead.regression",
"EsmClassificationHead.dense",
"EsmClassificationHead.out_proj",
]
)
mannequin = get_peft_model(mannequin, peft_config)Submit a SageMaker coaching job
After you might have outlined your coaching script, you may configure and submit a SageMaker coaching job. First, specify the hyperparameters:
hyperparameters = {
"model_id": "fb/esm2_t33_650M_UR50D",
"epochs": 1,
"per_device_train_batch_size": 8,
"gradient_accumulation_steps": 4,
"use_gradient_checkpointing": True,
"lora": True,
}Subsequent, outline what metrics to seize from the coaching logs:
metric_definitions = [
{"Name": "epoch", "Regex": "'epoch': ([0-9.]*)"},
{
"Identify": "max_gpu_mem",
"Regex": "Max GPU reminiscence use throughout coaching: ([0-9.e-]*) MB",
},
{"Identify": "train_loss", "Regex": "'loss': ([0-9.e-]*)"},
{
"Identify": "train_samples_per_second",
"Regex": "'train_samples_per_second': ([0-9.e-]*)",
},
{"Identify": "eval_loss", "Regex": "'eval_loss': ([0-9.e-]*)"},
{"Identify": "eval_accuracy", "Regex": "'eval_accuracy': ([0-9.e-]*)"},
]Lastly, outline a Hugging Face estimator and submit it for coaching on an ml.g5.2xlarge occasion kind. This can be a cost-effective occasion kind that’s broadly accessible in lots of AWS Areas:
from sagemaker.experiments.run import Run
from sagemaker.huggingface import HuggingFace
from sagemaker.inputs import TrainingInput
hf_estimator = HuggingFace(
base_job_name="esm-2-membrane-ft",
entry_point="lora-train.py",
source_dir="scripts",
instance_type="ml.g5.2xlarge",
instance_count=1,
transformers_version="4.28",
pytorch_version="2.0",
py_version="py310",
output_path=f"{S3_PATH}/output",
function=sagemaker_execution_role,
hyperparameters=hyperparameters,
metric_definitions=metric_definitions,
checkpoint_local_path="/choose/ml/checkpoints",
sagemaker_session=sagemaker_session,
keep_alive_period_in_seconds=3600,
tags=[{"Key": "project", "Value": "esm-fine-tuning"}],
)
with Run(
experiment_name=EXPERIMENT_NAME,
sagemaker_session=sagemaker_session,
) as run:
hf_estimator.match(
{
"prepare": TrainingInput(s3_data=train_s3_uri),
"take a look at": TrainingInput(s3_data=test_s3_uri),
}
)The next desk compares the completely different coaching strategies we mentioned and their impact on the runtime, accuracy, and GPU reminiscence necessities of our job.
| Configuration | Billable Time (min) | Analysis Accuracy | Max GPU Reminiscence Utilization (GB) |
| Base Mannequin | 28 | 0.91 | 22.6 |
| Base + GA | 21 | 0.90 | 17.8 |
| Base + GC | 29 | 0.91 | 10.2 |
| Base + LoRA | 23 | 0.90 | 18.6 |
The entire strategies produced fashions with excessive analysis accuracy. Utilizing LoRA and gradient activation decreased the runtime (and value) by 18% and 25%, respectively. Utilizing gradient checkpointing decreased the utmost GPU reminiscence utilization by 55%. Relying in your constraints (value, time, {hardware}), one among these approaches might make extra sense than one other.
Every of those strategies carry out effectively by themselves, however what occurs once we use them together? The next desk summarizes the outcomes.
| Configuration | Billable Time (min) | Analysis Accuracy | Max GPU Reminiscence Utilization (GB) |
| All strategies | 12 | 0.80 | 3.3 |
On this case, we see a 12% discount in accuracy. Nevertheless, we’ve decreased the runtime by 57% and GPU reminiscence use by 85%! This can be a large lower that enables us to coach on a variety of cost-effective occasion varieties.
Clear up
For those who’re following alongside in your personal AWS account, delete the any real-time inference endpoints and information you created to keep away from additional expenses.
predictor.delete_endpoint()
bucket = boto_session.useful resource("s3").Bucket(S3_BUCKET)
bucket.objects.filter(Prefix=S3_PREFIX).delete()Conclusion
On this publish, we demonstrated the way to effectively fine-tune protein language fashions like ESM-2 for a scientifically related process. For extra details about utilizing the Transformers and PEFT libraries to coach pLMS, take a look at the posts Deep Learning With Proteins and ESMBind (ESMB): Low Rank Adaptation of ESM-2 for Protein Binding Site Prediction on the Hugging Face weblog. You may as well discover extra examples of utilizing machine studying to foretell protein properties within the Awesome Protein Analysis on AWS GitHub repository.
Concerning the Writer
 Brian Loyal is a Senior AI/ML Options Architect within the International Healthcare and Life Sciences workforce at Amazon Net Providers. He has greater than 17 years’ expertise in biotechnology and machine studying, and is enthusiastic about serving to prospects resolve genomic and proteomic challenges. In his spare time, he enjoys cooking and consuming together with his family and friends.
Brian Loyal is a Senior AI/ML Options Architect within the International Healthcare and Life Sciences workforce at Amazon Net Providers. He has greater than 17 years’ expertise in biotechnology and machine studying, and is enthusiastic about serving to prospects resolve genomic and proteomic challenges. In his spare time, he enjoys cooking and consuming together with his family and friends.