Meet SPACEL: A New Deep-Studying-based Evaluation Toolkit for Spatial Transcriptomics
Scientists historically study tissues by analyzing the expression ranges of genes in particular person cells utilizing a method generally known as spatial transcriptomics (ST). Researchers acquire insights into cells’ spatial group and performance by measuring the amount of RNA in particular places inside a tissue. Spatial transcriptomics (ST) applied sciences have been instrumental in unraveling the mysteries of mRNA expression in particular person cells whereas sustaining their spatial coordinates. Nonetheless, challenges come up when a number of tissue slices have to be collectively analyzed, and the dimensions of spots in ST slices hampers the decision.
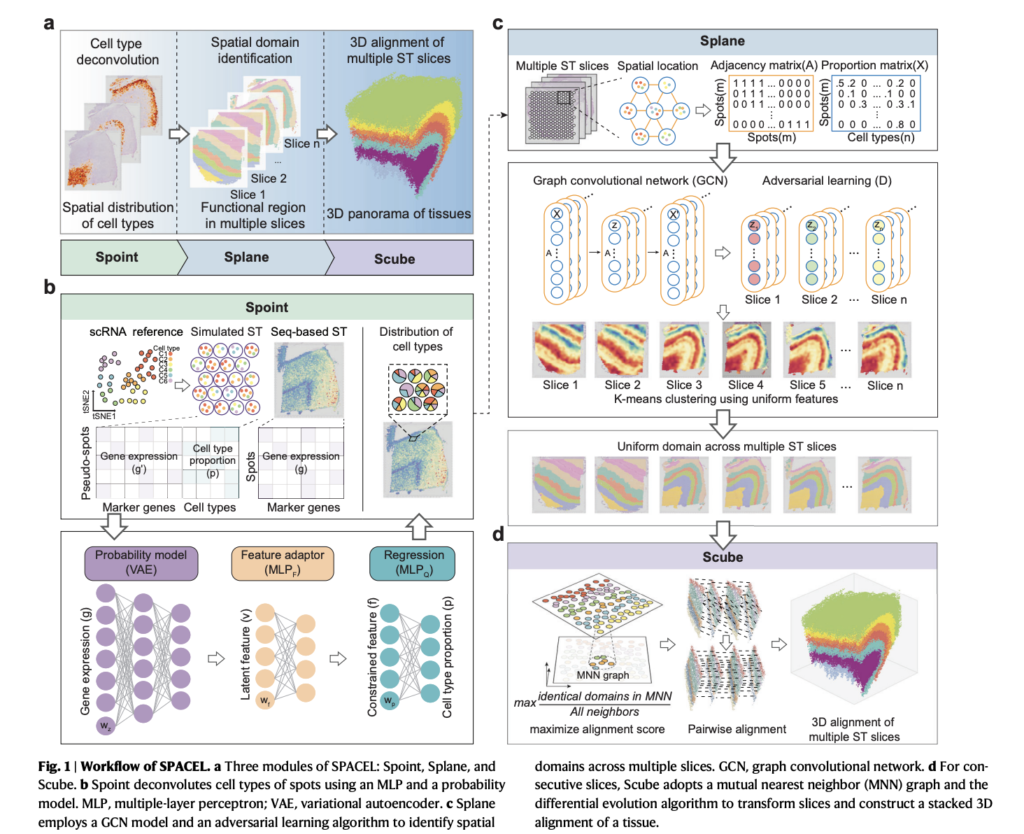
To beat these limitations, a bunch of researchers headed by Prof. Qu Kun from the College of Science and Expertise of the Chinese language Academy of Sciences has created an answer referred to as Spatial Structure Characterization by Deep Studying (SPACEL). This toolkit has three modules—Spoint, Splane, and Scube—that mix to create a 3D panorama of tissues routinely.
The primary module, Dash, tackles the cell-type deconvolution job. It predicts the spatial distribution of cell varieties utilizing a mix of simulated pseudo-spots, neural community modeling, and statistical restoration of expression profiles. This makes predictions correct and highly effective. The second module, Splane, makes use of a graph convolutional community (GCN) method and an adversarial studying algorithm to establish particular domains by collectively analyzing a number of ST slices. Splane makes use of adversarial coaching to take away batch results over a number of slices and makes use of cell-type composition as enter. Splane stands out for its progressive methodology of effectively figuring out spatial domains. The third module, Scube, automates the alignment of slices and constructs a stacked 3D structure of the tissue. That is essential in overcoming the challenges posed by the restrictions of experimental ST methods, permitting for a complete understanding of the tissue’s three-dimensional construction.
The researchers utilized SPACEL to 11 ST datasets totaling 156 slices and utilized applied sciences like 10X Visium, STARmap, MERFISH, Stereo-seq, and Spatial Transcriptomics. The researchers emphasize that SPACEL outperformed earlier methods in three basic analytical duties—cell sort distribution prediction, spatial area identification, and three-dimensional tissue reconstruction.
Additional, SPACEL demonstrated its superiority in cell sort deconvolution, spatial area identification, and 3D alignment towards 19 cutting-edge methods on simulated and actual ST datasets, with its superior efficiency over earlier methods and simplified method to precisely understanding ST knowledge.
In conclusion, SPACEL’s introduction is a big step in spatial transcriptomics. Its three modules present researchers with a robust instrument to beat the challenges related to joint evaluation of a number of ST slices, enabling exact cell sort predictions, efficient spatial area identification, and correct 3D tissue alignment. This instrument permits for correct 3D tissue alignment, cell sort predictions, and environment friendly spatial area identification.
Try the Paper. All credit score for this analysis goes to the researchers of this venture. Additionally, don’t neglect to affix our 35k+ ML SubReddit, 41k+ Facebook Community, Discord Channel, LinkedIn Group, Twitter, and Email Newsletter, the place we share the newest AI analysis information, cool AI tasks, and extra.
If you like our work, you will love our newsletter..
Rachit Ranjan is a consulting intern at MarktechPost . He’s at the moment pursuing his B.Tech from Indian Institute of Expertise(IIT) Patna . He’s actively shaping his profession within the subject of Synthetic Intelligence and Knowledge Science and is passionate and devoted for exploring these fields.