Meet PepCNN: A Deep Studying Device for Predicting Peptide Binding Residues in Proteins Utilizing Sequence, Structural, and Language Mannequin Options
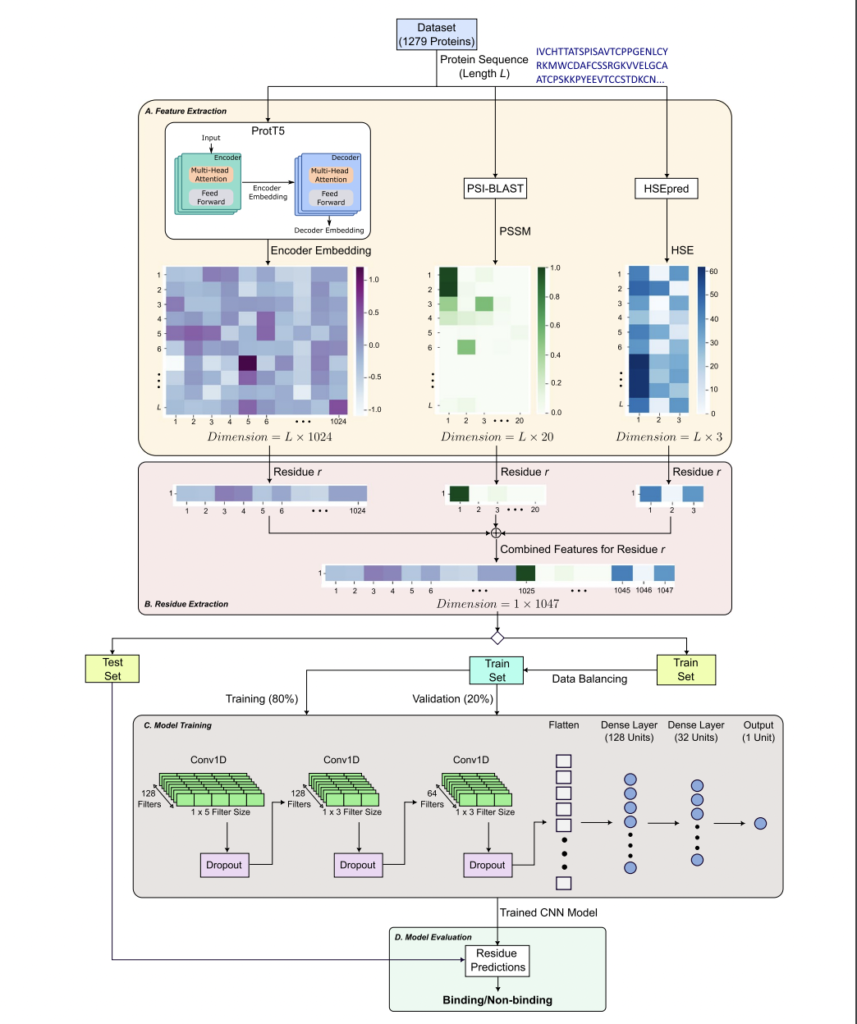
PepCNN, a deep studying mannequin developed by researchers from Griffith College, RIKEN Heart for Integrative Medical Sciences, Rutgers College, and The College of Tokyo, addresses the issue of predicting protein-peptide binding residues. PepCNN outperforms different strategies when it comes to specificity, precision, and AUC metrics by combining structural and sequence-based info, making it a invaluable instrument for understanding protein-peptide interactions and advancing drug discovery efforts.
Understanding protein-peptide interactions is essential for mobile processes and illness mechanisms like most cancers, necessitating computational strategies as experimental approaches are resource-intensive. Computational fashions, categorized into structure-based and sequence-based, provide options. Using options from pre-trained protein language fashions and publicity knowledge, PepCNN outperforms earlier strategies, emphasizing the importance of its characteristic set for improved prediction accuracy in protein-peptide interactions.
There’s a want for computational approaches to realize a deeper understanding of protein-peptide interactions and their position in mobile processes and illness mechanisms. Whereas structure-based and sequence-based fashions have been developed, accuracy stays a problem because of the complexity of the interactions. PepCNN, a novel deep studying mannequin, has been proposed to unravel this problem by integrating structural and sequence-based info to foretell peptide binding residues. With superior efficiency in comparison with present strategies, PepCNN is a promising instrument for supporting drug discovery efforts and advancing the understanding of protein-peptide interactions.
PepCNN makes use of progressive strategies equivalent to half-sphere publicity, position-specific scoring matrices, and embedding from a pre-trained protein language mannequin to attain superior outcomes in comparison with 9 present strategies, together with PepBCL. Its spectacular specificity and precision stand out, and its efficiency surpasses different state-of-the-art strategies. These developments spotlight the effectiveness of the proposed methodology.
The deep studying prediction mannequin, PepCNN, outperformed varied strategies, together with PepBCL, with increased specificity, precision, and AUC. After being evaluated on two take a look at units, PepCNN displayed notable enhancements, notably in AUC. The outcomes confirmed that sensitivity was 0.254, specificity was 0.988, precision was 0.55, MCC was 0.350, and AUC was 0.843 on the primary take a look at set. Future analysis goals to combine DeepInsight expertise to facilitate the appliance of 2D CNN architectures and switch studying strategies for additional developments.
In conclusion, the superior deep-learning prediction mannequin, PepCNN, incorporating structural and sequence-based info from major protein sequences, outperforms present strategies in specificity, precision, and AUC, as demonstrated in checks performed on TE125 and TE639 datasets. Additional analysis goals to boost its efficiency by integrating DeepInsight expertise, enabling the appliance of 2D CNN architectures and switch studying strategies.
Take a look at the Paper and Github. All credit score for this analysis goes to the researchers of this venture. Additionally, don’t neglect to affix our 33k+ ML SubReddit, 41k+ Facebook Community, Discord Channel, and Email Newsletter, the place we share the most recent AI analysis information, cool AI initiatives, and extra.
If you like our work, you will love our newsletter..
Sana Hassan, a consulting intern at Marktechpost and dual-degree scholar at IIT Madras, is captivated with making use of expertise and AI to deal with real-world challenges. With a eager curiosity in fixing sensible issues, he brings a recent perspective to the intersection of AI and real-life options.